Systematics
Integrative taxonomy
Researchers: Dr. Thomas Lecocq, Nicolas Brasero, Prof. Denis Michez, Prof. Pierre Rasmont
Species are one of the fundamental units in biology and accurate species delimitation studies is essential for assessing taxonomic status of biodiversity, conservation or biological control projects. Nevertheless, criteria for delimiting species and subspecies is still confusing and controversial. Numerous methods of species delimitation have been proposed but there is no universal consensus. The current taxonomy, especially for invertebrate groups, is mainly based on morphological approach criteria. However, identifying closely related species is often hindered by lack of diagnostic morphological characters (i.e. cryptic species). Integrative taxonomy combine multiple and complementary perspectives (phylogeography, comparative morphology, eco-chemical traits, population genetics, ecology, development, behavior, etc.) in order to delineate species boundary.
Our research compares taxonomic conclusions obtained from the commonly used lines of evidence for species delimitation in bumblebees (geometric morphometric of wing shape, genetic differentiation assessment, sequence-based species delimitation methods, and differentiation of cephalic labial gland secretions). We ultimately aim to assess the usefulness of these lines of evidence as components of an integrative decision framework to delimit bumblebee species. Our results show that analyses based on wing shape do not delineate any obvious cluster. In contrast, nuclear/mitochondrial, sequence-based species delimitation methods, and analyses based on cephalic labial gland secretions are congruent with each other. This allows setting up an integrative decision framework to establish strongly supported species and subspecies status within bumblebees.
Selected publications:
- Lecocq, T., Brasero, N., De Meulemeester, T., Michez, D., Dellicour, S., Lhomme, P., de Jonghe, R., Valterová, I., Urbanová, K., Rasmont, P., 2014. An integrative taxonomic approach to assess the status of Corsican bumblebees: implications for conservation. Anim. Conserv. n/a–n/a. doi:10.1111/acv.12164
- Lecocq, T., Dellicour, S., Michez, D., Dehon, M., Dewulf, A., De Meulemeester, T., Brasero, N., Valterová, I., Rasplus, J.-Y., Rasmont, P., 2015. Methods for species delimitation in bumblebees (Hymenoptera, Apidae, Bombus): towards an integrative approach. Zool. Scr.
- Lecocq, T., Lhomme, P., Michez, D., Dellicour, S., Valterová, I., Rasmont, P., 2011. Molecular and chemical characters to evaluate species status of two cuckoo bumblebees: Bombus barbutellus and Bombus maxillosus (Hymenoptera, Apidae, Bombini). Syst. Entomol. 36, 453–469. doi:10.1111/j.1365-3113.2011.00576.x
Paleontology
Researchers: Dr. Denis Michez, Manuel Dehon
Bee fossils are relatively rare in deposits. A bit less than 200 fossil species were described so far. Because bee fossils are scarce, additional records are therefore of significant interest to evaluate the origin of clades, re-evaluate hypotheses about their phylogenetic relationships, document patterns of distribution, ascertain paleobiological associations and ancient behaviors, estimate rates of diversification/extinction, and calibrate accurately molecular phylogenies.
We recently developed in the laboratory of Zoology an important data set of wing shape and a core of methods based on geometric morphometrics to define phenetic affinities between extant and extinct clades of bees. This methodology was particularly efficient for compression fossils whereby wings are often better preserved than other body parts.
We are now developing new dataset to revise all available bee fossils. Moreover we will use this taxonomically revised fossils to propose re-calibration of molecular phylogenies of some key taxa.
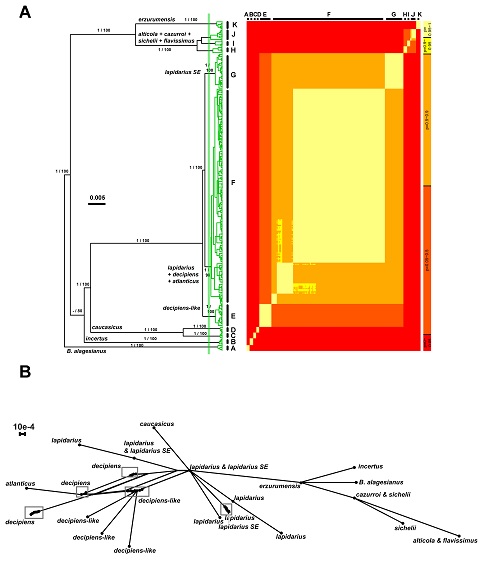
Phylogenetic, GMYC and bGMYC analyses. A. Bayesian ultrametric tree based on COI sequences with single threshold GMYC model applied and bGMYC pairwise probability of conspecificity. Values above tree branches are Bayesian posterior probabilities / maximum likelihood bootstrap values. The green branches are entities detected with the single-threshold GMYC method. The adjusted single threshold from the GMYC model is shown by the vertical green bare. The colored matrix corresponds to the pairwise probabilities of conspecificity returned by the bGMYC method (see also the related color scale on the right). B. Phylogenetic network based on PEPCK data matrix. The scale bar represents the split support for the edges. Dots are haplotypes. Names summarize all individuals from the same taxon included in one haplotype. Grey frames include several closely related haplotypes from the same taxon.
